
New Book on Cyber-Physical Distributed Systems
Wiley has published a new book on cyber-physical distributed systems by Huadong Mo, Giovanni Sansavini, and Min Xie.
The book presents recent advances in modeling, reliability analysis and applications of cyber-physical distributed systems. It provides an introduction to commonly used cyber-physical systems, and explains methods for stability analysis and optimal maintenance of these systems and integration of them to communications networks. The book is useful for both postgraduate and senior undergraduate courses in control systems, systems engineering and computer science.
Reference: H. Mo, G. Sansavini, and M. Xie, Cyber-Physical Distributed Systems: Modeling, Reliability Analysis and Applications, Wiley, 2021 (https://www.amazon.com/Cyber-Physical-Distributed-Systems-Reliability-Applications/dp/111968267, https://www.wiley.com/en-hk/Cyber+Physical+Distributed+Systems%3A+Modeling% 2C+Reliability+Analysis+and+Applications-p-97811196826773)

New Book on AI for Medicine and Life Science
Springer has published a new book Advances in Artificial Intelligence, Computation, and Data Science for Medicine and Life Science edited by Tuan Pham, Hong Yan, Muhammad Waqar Ashraf, and Folke Sjöberg.
The book presents latest development of AI, big data analytics, and computational algorithms, and their applications to biology, chemistry, physiology, and medicine. It has 15 chapters written by international experts. The book highlights the need of interdisciplinary collaboration and the application of new technologies to solving cutting-edge research problems. The topics covered in the book include biological networks, gene expression data analysis, drug development, disease diagnosis, medical imaging, biomechanics, and augment medicine.
Reference: T. Pham, H. Yan, M. W. Ashraf, and F. Sjöberg (Eds), Advances in Artificial Intelligence, Computation, and Data Science for Medicine and Life Science, Springer, 2021 (https://www.springer.com/gp/book/9783030699505).
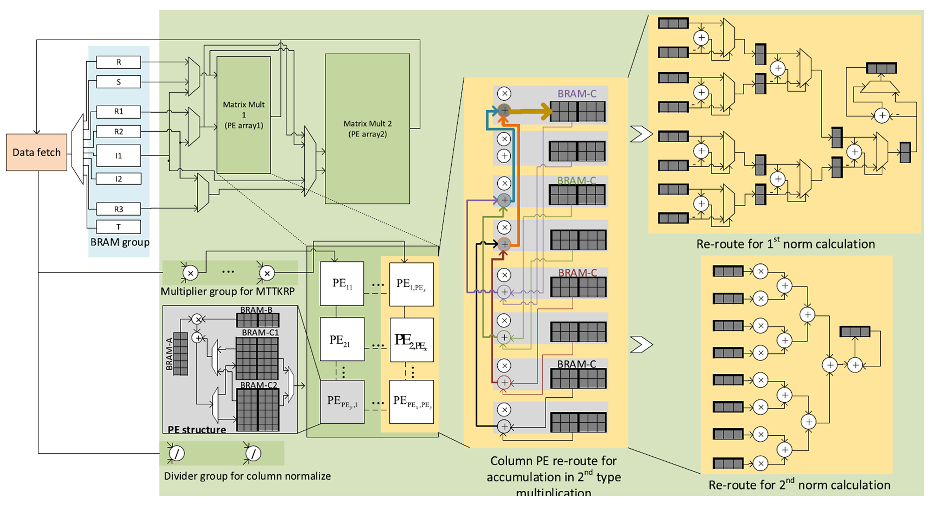
FPGA Based Parallel Processors for Dense Tensor Computation
A paper on FPGA for tensor computing Vic Weipei Huang, Ray Cheung and Hong Yan has been published in IEEE Transactions on Very Large Scale Integration (VLSI) Systems.
The team has developed a hardware architecture based on application specific instruction sets and a uniform processing element array to execute the instructions. They use the pipeline technology and resources sharing to shrink the data-path size of the processor. This processor cuts down the computing time by 50% compared with GPU and by 93% compared with an implementation on CPU.
The system is especially useful for tensor decomposition, which has many applications to signal processing, biomedical data analysis, and computer vision.
Reference: W.-P. Huang, R. C. C. Cheung, and H. Yan, “An efficient parallel processor for dense tensor computation," IEEE Transactions on Very Large Scale Integration (VLSI) Systems, 29(7):1335-1347, 2021 (https://ieeexplore.ieee.org/document/9442854).
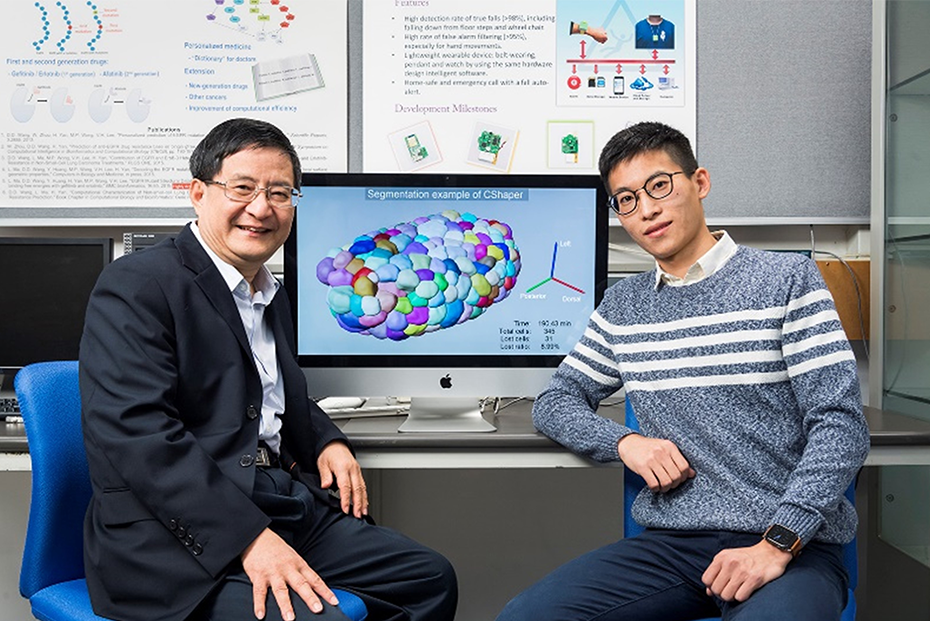
Analysis of 4D Cell Division Images
A computational framework called CShaper has been published in Nature Communications by Jianfeng Cao and Hong Yan in collaboration with biologists at Hong Kong Baptist University and Peking University.
According to Professor Yan, “Each animal (or human) is grown from a single cell, a fertilized egg. Amazingly, successive divisions of this cell and its daughter cells will form animal organs and body automatically. We use laser beams to obtain cell images at different depths and at different time points to form a 4D (xyzt) data array during cell division. Hong Kong Baptist University and Peking University did wet-lab experiments with C. elegans (worm) to produce terabytes of data, and CityU performed computational analysis. We developed machine learning methods to extract, track and visualize cells and analyse their functions.”
CShaper is a revolutionary tool for cell image analysis. It reduces cell annotation time to several hours using this tool, which would otherwise require several hundred hours if biologists do this manually. This technique can stimulate and enhance further studies in the fields of developmental biology, cell biology, and biomechanics.
Reference: J. Cao, G. Guan, V. W. S. Ho, M. K. Wong, L. Y. Chan, C.Tang, Z. Y. Zhao, and H. Yan, "Establishment of morphological atlas of Caenorhabditis elegans embryo using deep-learning-based 4D segmentation," Nature Communications, 11:6254:1-14, 2020 (https://www.nature.com/articles/s41467-020-19863-x).
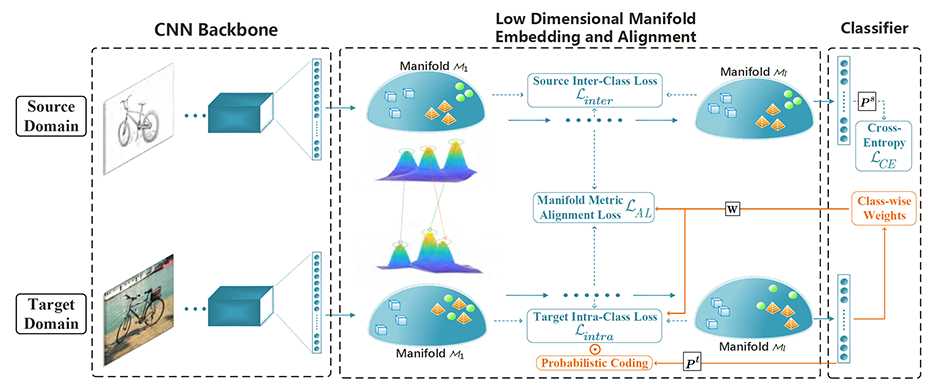
Unsupervised Domain Adaptation in Deep Learning
A paper on domain adaptation has been published in IEEE Transactions on Pattern Analysis and Machine Intelligence (PAMI) by You-Wei Luo, Chuan-Xian Ren, Dao-Qing Dai, and Hong Yan.
Deep learning has found many applications in artificial intelligence, pattern recognition and big data analysis. However, the performance of a deep network often depends on the availability of a large number of high-quality training samples. The collect of these samples can be highly labor intensive and costly in real-world applications. This paper solves the problem by using a strategy called unsupervised domain adaptation (UDA). It “is designed to deal with the shortage of labels by leveraging rich labels and strong supervision from the source domain to the target domain where there is no access to the annotations”. The authors have proposed “a discriminative manifold propagation method for both vanilla and partial UDA problems”. The method is verified to be effective through many data classification experiments.
Reference: Y. W. Luo, C. X. Ren, D. Q. Dai, and H. Yan, "Unsupervised domain adaptation via discriminative manifold propagation," IEEE Transactions on Pattern Analysis and Machine Intelligence, early access published in 2020 (https://ieeexplore.ieee.org/document/9158545).

Tensor Decomposition Based on Random Projections
Recently Maolin Che, Yimin Wei, and Hong Yan have published an efficient method for computing tensor decomposition in SIMAX.
Tensor data often have low ranks. That is, a large multidimensional data array can be approximated by a much smaller array. A method for doing this is to compute tensor decomposition. In this paper, the authors propose an algorithm for singular value decomposition (SVD) of high order tensors using the power iteration scheme and random projections. The proposed technique has a favorable performance compared with existing ones. The authors have also provided a rigorous mathematical analysis of the error bound of the algorithm.
This method is useful for big data analysis, for example, to reduce the dimensionality of the data in machine learning and pattern classification.
Reference: M. Che, Y, Wei, and H, Yan, “The computation of low multilinear rank approximations of tensors via power scheme and random projection,” SIAM Journal on Matrix Analysis and Applications (SIMAX), 41(2):605-636, 2020 (https://epubs.siam.org/doi/abs/10.1137/19M1237016).
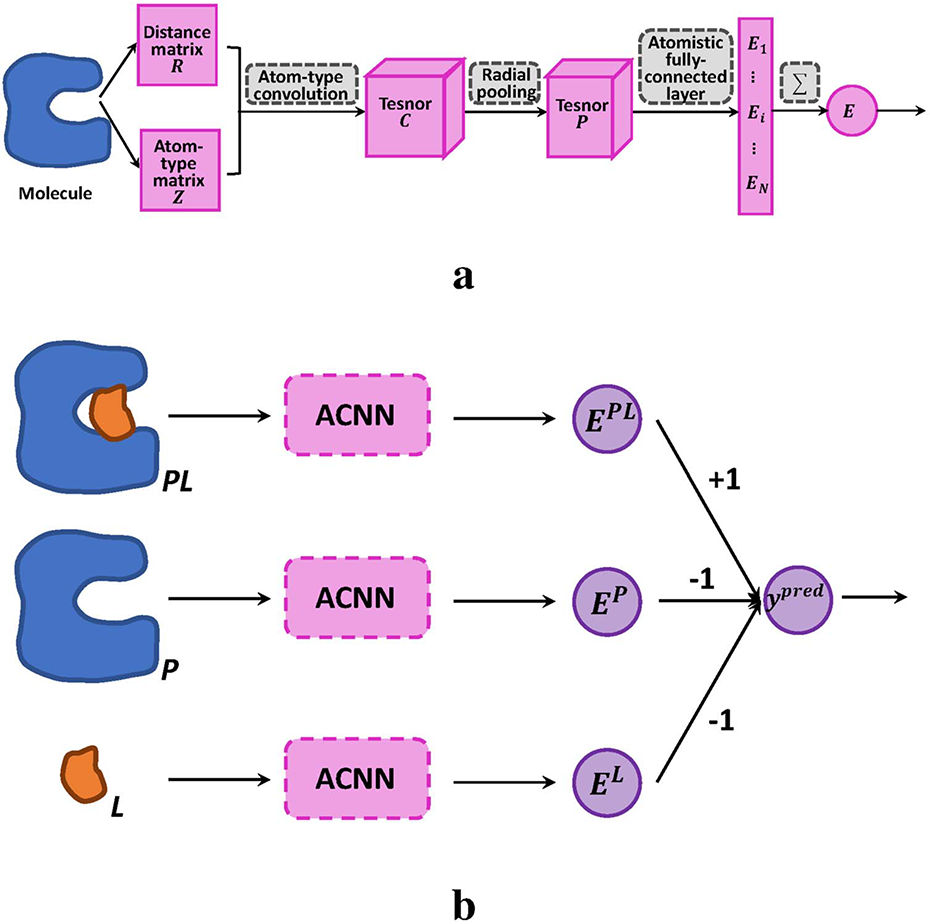
Scoring Functions for Biomolecular Modeling and Analysis
Debby Wang, Mengxu Zhu and Hong Yan have recently published a paper on scoring functions of biomolecular complexes in Briefings in Bioinformatics.
It is important to study biomolecular interactions to design new drugs and assess the therapeutic efficacies of existing ones. For example, drug resistance arises due to a protein mutation that causes the reduction of protein-drug binding strength. Such strength can be measured mathematically using a scoring function. In this paper, the authors compare two approaches: free energy-based simulations and machine learning-based scoring functions. They presented several case studies, and discussed the strengths and weaknesses of the two types of methods.
This work provides a useful guide for the selection of scoring functions and points out the areas of potential improvement to formulate more robust functions in biomolecular modeling and analysis.
Reference: D. D. Wang, M. Zhu, and H. Yan, “Computationally predicting binding affinity in protein–ligand complexes: free energy-based simulations and machine learning-based scoring functions,” Briefings in Bioinformatics, published online in 2020 (https://academic.oup.com/bib/article-abstract/22/3/bbaa107/5860693).
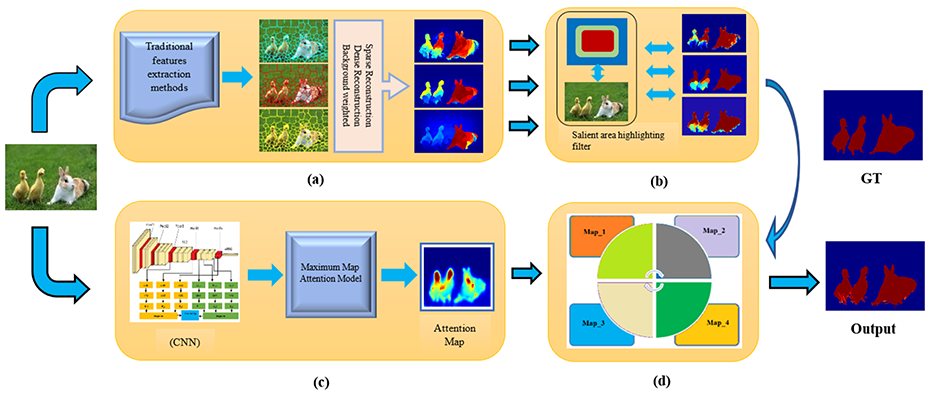
Saliency Detection Using Deep Features
Mehmood Nawaz and Hong Yan have published a paper on saliency detection using deep features in IEEE Transactions on Multimedia.
Commonly used saliency detection methods rely on contrast between background and foreground or contour information. They can be sensitive to noise. In this paper, the authors compute the contrast of image based on both supervised and unsupervised learning. They have developed an affinity-based robust background subtraction algorithm to construct the maximum attention map. The method works well for images with noisy and cluttered backgrounds.
Figure and background separation is often the first step in object recognition. The saliency detection method proposed in this paper has useful applications to many computer vision systems for object detection and classification.
Reference: M. Nawaz and H. Yan, “Saliency detection using deep features and affinity-based robust background subtraction,” IEEE Transactions on Multimedia, early access published in 2020 (https://ieeexplore.ieee.org/document/9180092).
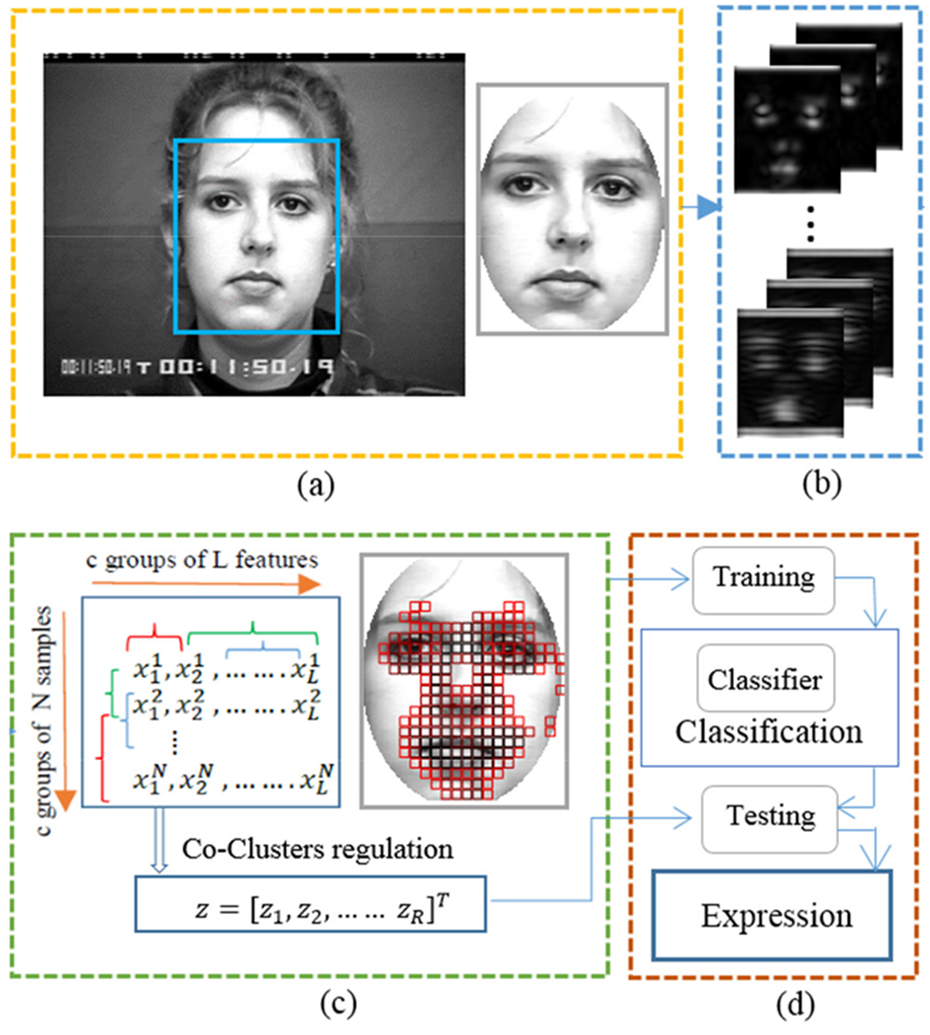
Co-clustering Based Feature Selection in Facial Expression Recognition
Sheheryar Khan, Lijiang Chen, and Hong Yan have published a paper on co-clustering based feature selection method for human facial expression recognition in IEEE Transaction on Affective Computing.
Gabor wavelets are proven to be robust features for human facial expression. However, there can be tens of thousands of features due to many scales and orientations. This paper presents a robust method for feature selection based on co-clustering. In commonly used clustering methods, we group data in either feature or sample direction, but not both. In co-clustering, we do the grouping in both directions. This is naturally more complicated and is often considered intractable. Our group developed and patented a hyperplane based co-clustering method. This method is further improved using an SVD based algorithm. In this paper, co-clustering is employed to extract useful features for facial expression recognition. The technique can still obtain an accuracy over 90% when we use as little as 0.9% of the original Gabor wavelet data.
It’s the first time that co-clustering is used for feature selection, which is a significant problem in pattern recognition. It is interesting that the features extracted automatically by our algorithm are consistent with the Action Units (AUs) manually identified by other researchers.
Reference: S. Khan, L. Chen, and H. Yan, “Co-clustering to reveal salient facial features for expression recognition,” IEEE Transactions on Affective Computing, 11(2):348-360, 2020 (https://ieeexplore.ieee.org/document/8186192).
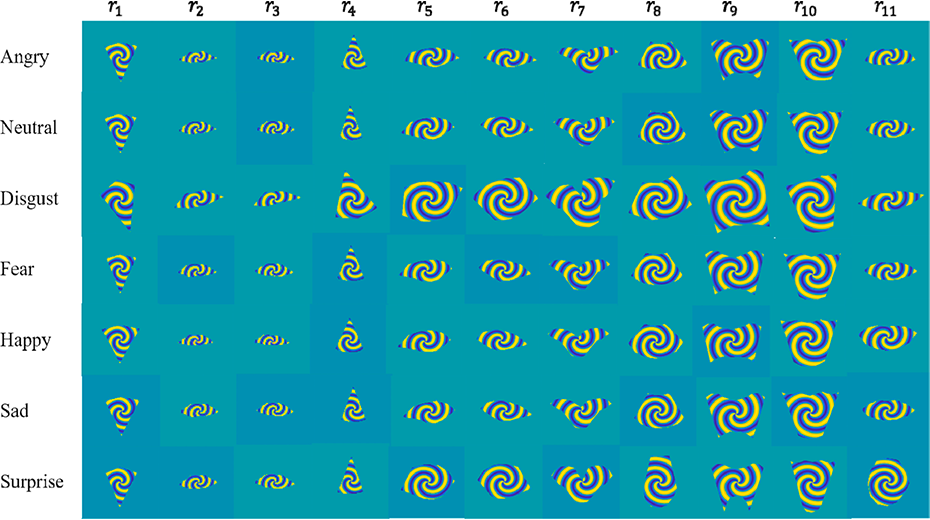
Contour and Region Harmonics for Human Facial Expression Recognition
Ali Raza Shahid, Sheheryar Khan, and Hong Yan have published a paper on human facial expression analysis in Journal of Visual Communication and Image Representation.
The team has discovered that contour and region harmonics from the human face provide powerful features for facial expression recognition. The movements of facial regions can be considered as time series and Fourier coefficients of these time series show distinct patterns in the radial and angular frequency domain. These features can be used to build a robust classifier. The proposed technique is demonstrated to perform better in terms of computing time and classification accuracy than existing methods through many experiments on several facial expression databases.
Facial expression recognition has many applications in human-machine interaction systems, mobile computing, electronic games, and multimedia and digital entertainment systems.
Reference: A. R. Shahid, S. Khan, and H. Yan, "Contour and region harmonic features for sub-local facial expression recognition," Journal of Visual Communication and Image Representation, 73:102949, 2020 (https://www.sciencedirect.com/ science/article/abs/pii/S1047320320301772).
Recent Top Conference Publications
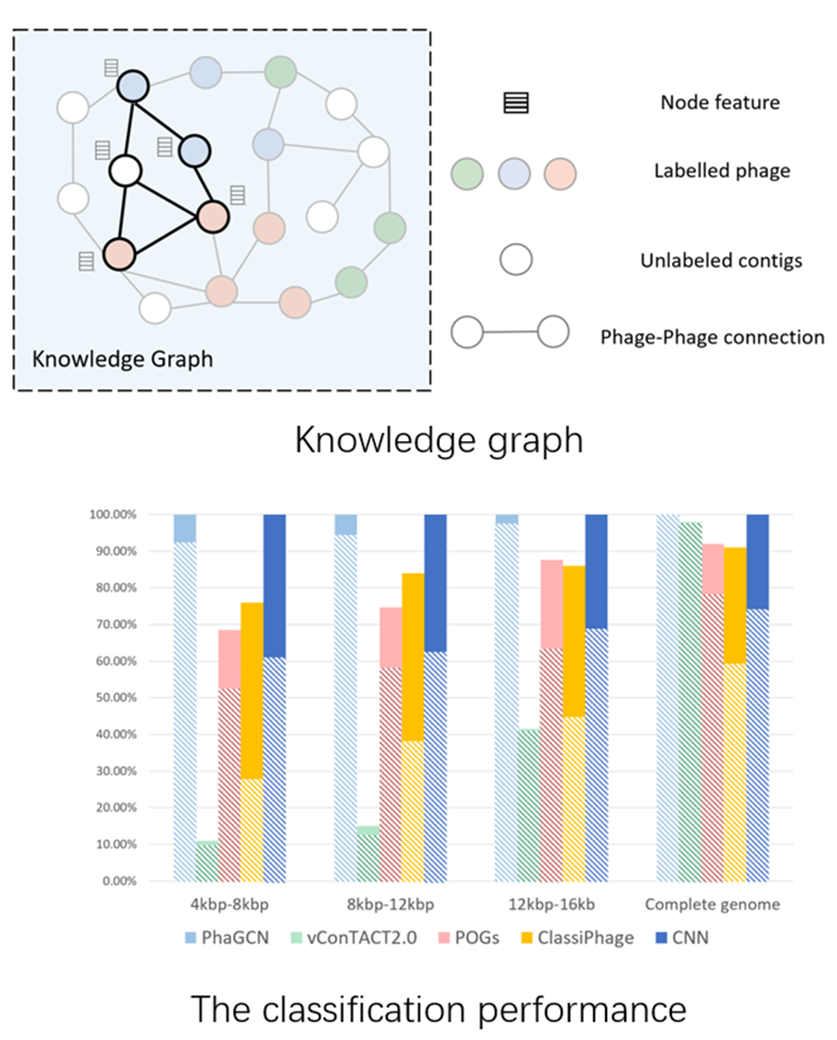
Bacteriophage Classification using CNN
Dr Yanni Sun and her Ph.D. student Mr. Jiayu Shang recently published a paper Bacteriophage classification for assembled contigs using graph convolutional network at ISMB/ECCB 2021, a top conference in the field of computational biology and bioinformatics, which has an acceptance rate of only 18.6%
Bacteriophages (or phages) are a type of virus. However, rather than infecting human/animals, they infect and kill bacteria. Thus, phages have potential to be used as antibiotics, which can help us fight the superbug (i.e., bacteria resistant to antibiotics). A fundamental need to use phages as antibiotics is to find them and label them. However, phages are the most abundant and diverse biological entities and characterized phages are just the tip of the iceberg. The goal of this project is to automatically and accurately mine new phages and assign their taxonomic groups from DNA sequencing data. The main challenge is there are only limited known phages (few training data).
In this work, the authors present a novel semi-supervised learning model, named PhaGCN, to conduct taxonomic classification for phage contigs. In this learning model, they construct a knowledge graph by combining the DNA sequence features learned by convolutional neural network and protein sequence similarity gained from gene-sharing network. Then they apply graph convolutional network to utilize both the labeled and unlabeled samples in training to enhance the learning ability.
.png)
Federated Learning Framework for Medical Image Analysis
Jie Liu, Xiaoqing Guo, and Yixuan Yuan proposed a novel federated learning framework for lesion classification to protect the privacy and ethical concerns in medical image analysis. Their paper “Prototypical interaction graph for unsupervised domain adaptation in surgical instrument segmentation” has been accepted by MICCAI 2021, the 24th International Conference on Medical Image Computing and Computer Assisted Intervention. Jie Liu received MICCAI Student Travel Award.
.png)
Unsupervised Domain Adaptation in Semantic Segmentation
Xiaoqing Guo, Chen Yang, Baopu Li, and Yixuan Yuan have proposed a new MetaCorrection framework to address unsupervised domain adaptation (UDA), which aims to transfer the knowledge from the labeled source domain to the unlabeled target domain in object segmentation. This work was presented at CVPR 2021.
Go to top

